The Multimodal Algorithms for Health Deep Dive
This blog post provides a recent exploration into multimodal models related to health. Especially in the case of diagnosis, we hardly ever rely on a single modality to inform our decisions, so we must rely on the multimodal nature of human beings to come up with a comprehensive view of what is going on. This won’t mean that we truly use “everything” in every paper, as in feature selection only specific modalities can inform us of an outcome, or the authors simply didn’t have enough data to dive deeper.
(2014)Fusion of multimodal medical images using Daubechies complex wavelet transform - A multiresolution approach
This first paper makes more sense given my previous post of wavelets and processing biosignals. This is another paper focusing on the concatenation of MRI and CT scans of the brain. This then means we’re dealing with an early fusion of medical images.
Wavelet transforms are a way of preprocessing and decomposing images into simpler components and representations. Here they denoise and enhance the image for fusion. The wavelet used here is a Daubechies complex wavelet transform applied to both CT and MRI images. Then a maximum selection rule is used for fusion such that noise is eliminated. There are better techniques that are used later and that were mentioned in my last blog being the (2019)A comparative review: Medical image fusion using SWT and DWT.
In the deep dives I do I try to cover good examples and give a breadth of information on what types of work is being done. This paper is mostly included to show that this field does exist and that these techniques are valid and give good results. They’re considered “multimodal”, but in my opinion doesn’t fit the
(2018)Deep Learning Role in Early Diagnosis of Prostate Cancer
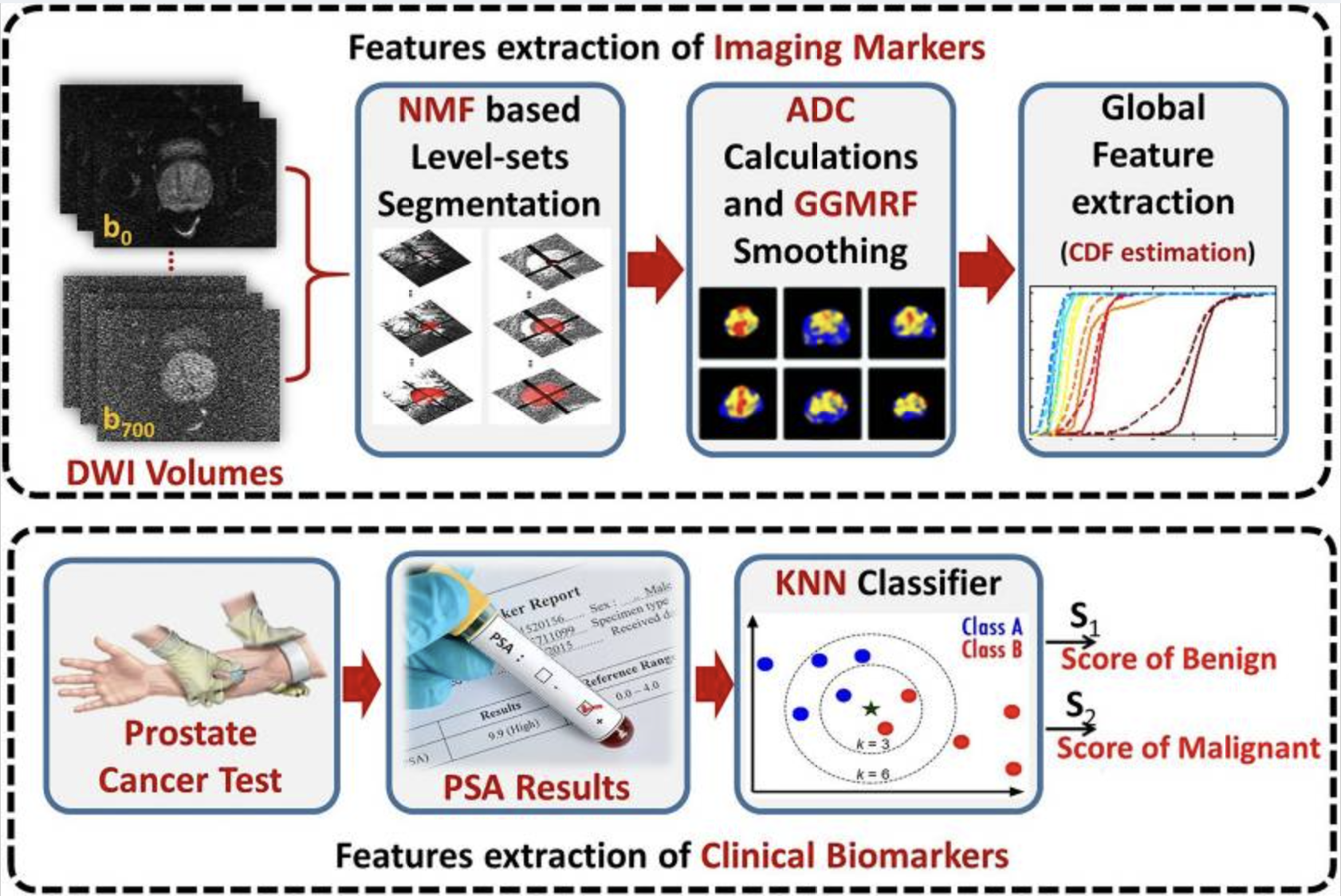
This was a super interesting paper on how a feed of a prostate camera is used to diagnose prostate cancer. Step one uses a level-set method (used in topology) for detecting the region of the prostate gland area. A nondeterministic speed function is used to guide the level set and then employs a 3D nonnegative matrix factorization (NMF) model. The NMF is used to determine how the level set boundary should evolve by taking in MRI intensity information, the prior topology, and spatial voxel interactions. The prediction is compared with the training data’s annotated ground truth.
After the level set boundary is changed, the DW-MRI image extracts the ADC (apparent diffusion coefficients). The ADC provides a measurement about the diffusion property of water molecules in tissues. These results are then normalized.
After normalization a smoothing takes place to replace the noise of the ADCs. A markov random field (MRF) is a model used to describe random neighboring pixels. This uses a generalized gaussian markov random field (GGMRF) meaning it takes into consideration a generalized gaussian distribution of random values.
A cumulative distribution function (CDF) is used to describe the distribution of ADCs. Any variation in CDF distributions can indicate prostate cancer. Lastly, we use an autoencoder, as they’re good in detecting rare events (colon cancer) in unbalanced datasets. They’re trained to reconstruct instances where the data is similar, but when we show prostate cancer, the autoencoder will fail at reconstructing it (it’s never come across this in the training) and the error will be high. This way we can detect prostate cancer!
As for the clinical biomarkers, these are simply fed in with a KNN classifier. The probabilities for both the SNCSAE and KNN are assessed and a fusion SNSCAE gives a final diagnosis.
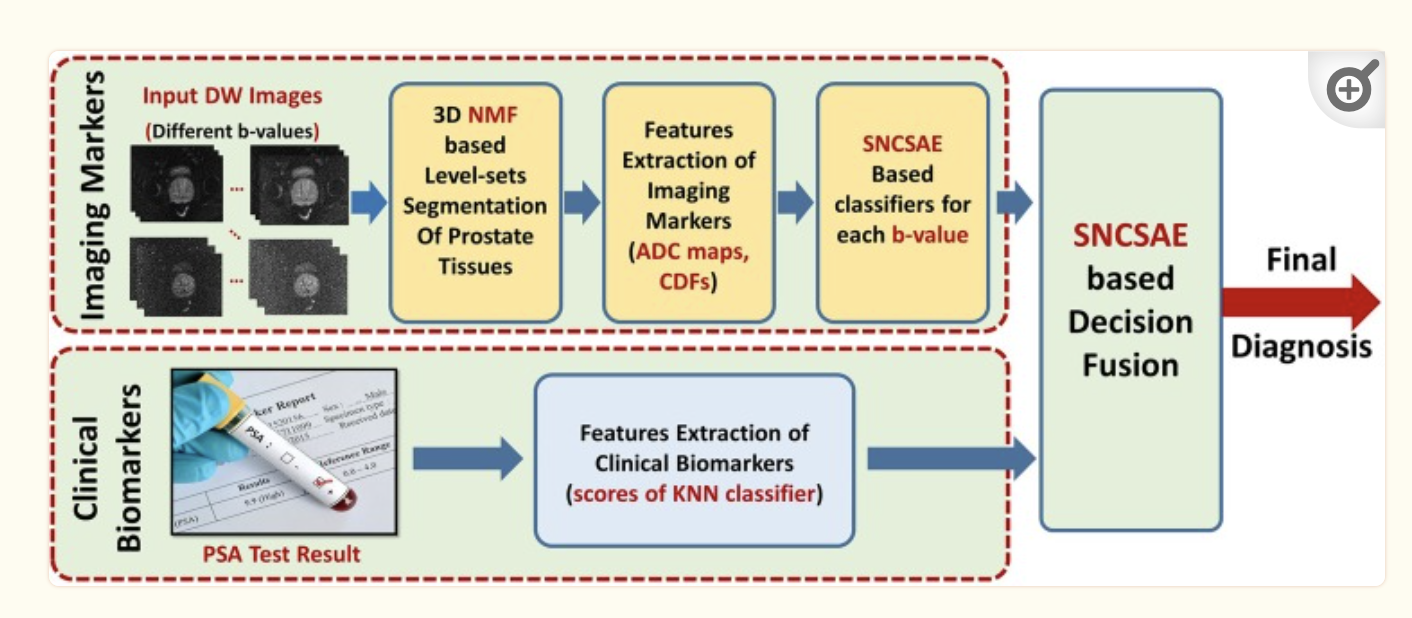
That was a lot! So let’s review in english. We get a feed of DW images that show us the prostate. The 3D NMF level-sets are used to find the region of interest. ADC maps are used to analyze that area of interest for the diffusion property of water molecules in tissues. These ADC measurements are normalized to make them easier to analyze with the GGMRF. A cumulative distribution function is used to describe the distribution of ADCs where steep portions of the CDF indicate a concentration of ADC values around a specific range as described by the level-set. Lastly an autoencoder that is trained on normal prostates will fail to recreate cancerous prostates as the CDFs will be different. The loss the autoencoder entails means that the algorithm has failed. Also, in this autoencoder are clinical biomarkers that are fed into a KNN classifier that help the autoencoder reach a final diagnosis.
(2018)Comparison of Machine-Learning Classification Models for Glaucoma Management
This paper has a really interesting way of determining multimodal features in that it uses the genetic algorithm. The problem lies in that a genetic algorithm doesn’t explore an efficient state of features, and we already have existing tests such as backward stepwise regression that’d probably do better in determining features.
Another huge issue with this paper is that there’s no outside testing set outside of the two sets fed into the genetic algorithm. That means this paper is incredibly susceptible to overfitting and shaky results.
(2018)A feature fusion system for basal cell carcinoma detection through data-driven feature learning and patient profile
Another intensely interesting paper here. BCC is a type of skin cancer that can only really show during its late stages. It is incredibly common with 4 million cases diagnosed in the US every year.
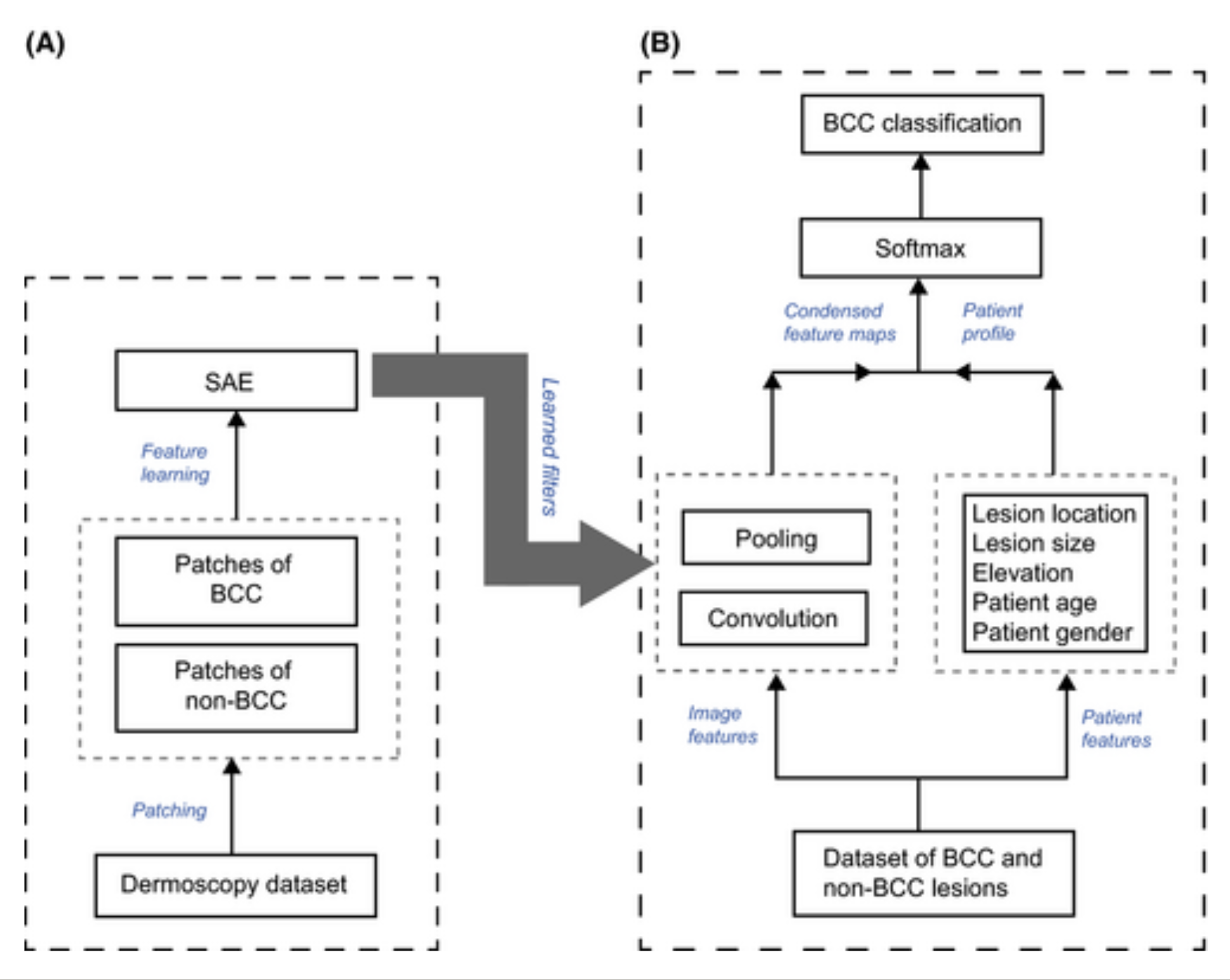
They use a sparse autoencoder (SAE) as their feature learning tool. Both BCC and non-BCC images are fed in. The perceptron weights of the SAE are treated as kernels used to convolve over each image. We’re essentially analyzing the activations of each weight on the new image fed into the autoencoder and convolving over those activations. We’re creating our own CNN by using an SAE as an initial kernel over the dataset of skin! This effectively specializes the kernel to understand skin patterns!
Pooling is used to reduce dimensionality of feature maps. The resulting feature map is combined with patient data for a softmax classifier resulting in a final classification.
(2018)Prediction of rupture risk in anterior communicating artery aneurysms with feed-forward artificial neural network
Anterior communicating artery (ACOM) aneurysms are the most common intracranial aneurysms, we’re looking to diagnose ruptured vs unruptured arteries. The dataset the authors had to work with has that of 594 ACOM aneurysms, 54 unruptured and 540 ruptured.
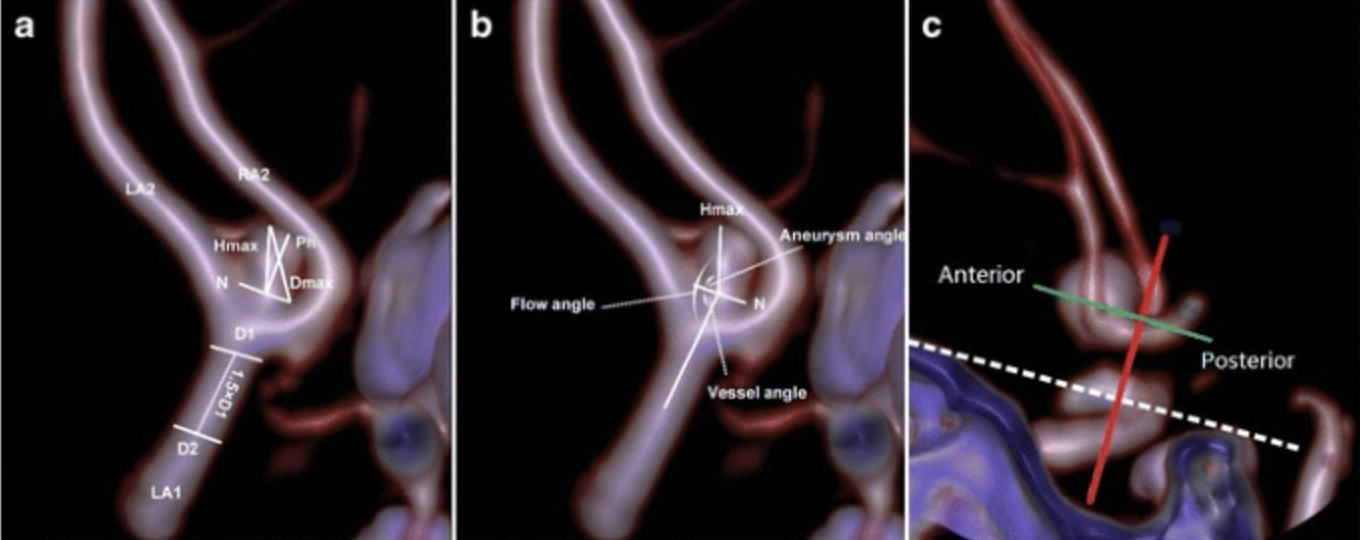
When collecting data an issue I saw was the lack of computer vision for aneurysms. Instead, the authors opted for hand-based measurements for each special part of the aneurysm. The problem being that any measurement has to be done by a professional who understands the structures which can differ drastically. This means that a professional diagnosis would’ve most likely been more accurate and faster in the first place that using this algorithm.
The inputs to a NN were then 17 parameters including the measurements and other values such as hypertension, smoking, age, etc.
An adaptive synthetic approach was used for the unbalanced dataset. Imagine we only have two samples in a given dataset. SMOTE is a technique that’ll make synthetic points on that line. ADASYN would then do the same, but move off the line slightly. Then just imagine how this expands for n dimensions and with many possible lines to choose from.
The results were good; 95% accurate. I’m just more concerned with how useful the work truly is. This paper is interesting and if you’re wondering “why even include this?” I had to ask myself more times than I would’ve liked with some other papers haha. I guess this is the best of what we’re given because there aren’t any objectively poor experiment design decisions unlike some other papers, this just suffers from practicality issues for me.
(2019)Multimodal Machine Learning-based Knee Osteoarthritis Progression Prediction from Plain Radiographs and Clinical Data
The authors develop a Knee osteoarthritis (OA) multimodal ML-based progression prediction model utilizing radiograph data, examination results and the previous medical history of patients.
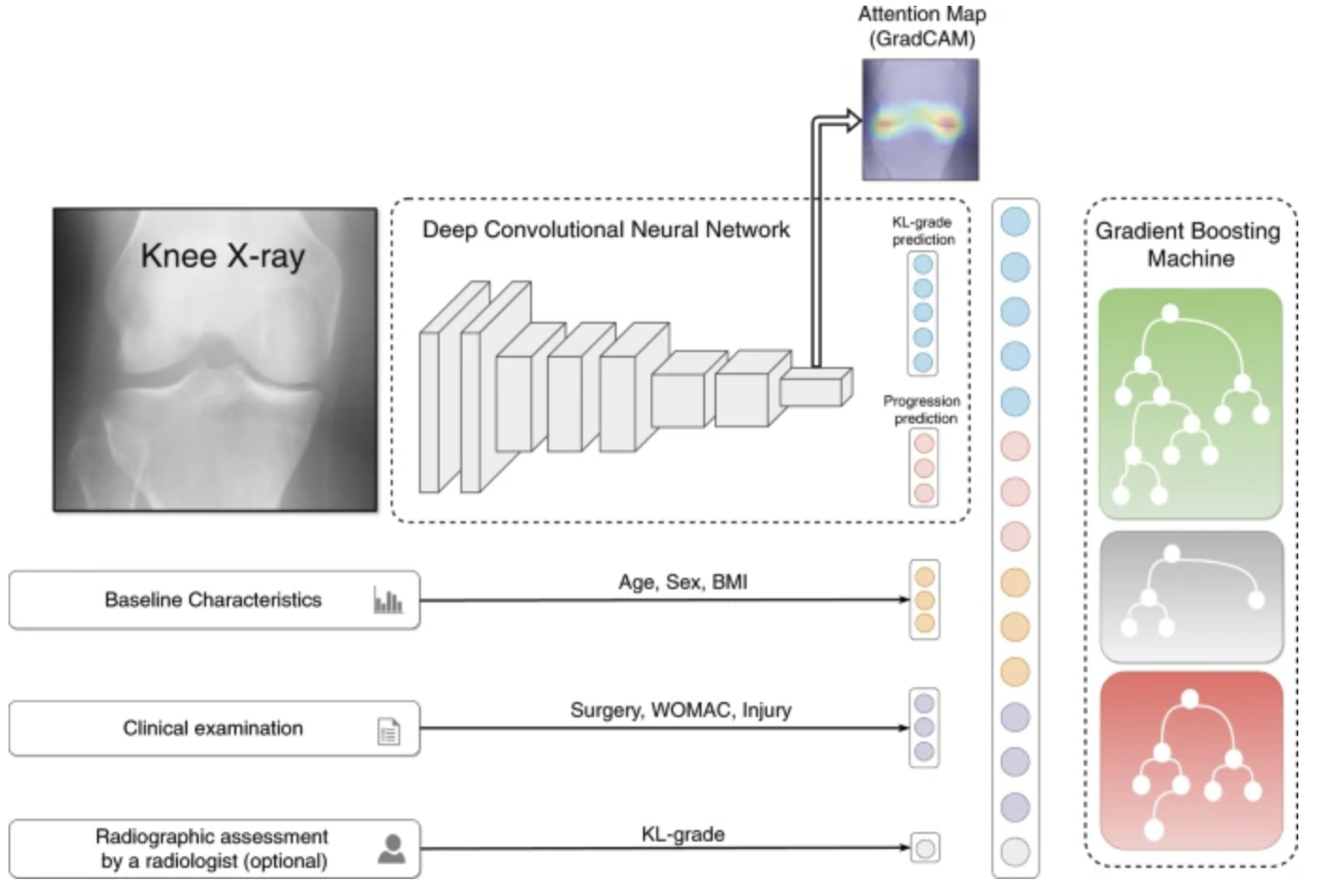
A deep CNN looks at knee x-rays initially. The unclear usage of GradCAM is basically just highlighting the activations for a doctor to see where the CNN is looking to come to its conclusions. We get a final prediction of where the knee is in its deterioration as well as a prediction of where it’ll go from the CNN. This embedding is then concatenated with other EHR data such as Age, Surgery information, and the optional radiographic assessment. This last assessment may do some heavy lifting in terms of the predictive power and I’m afraid it may skew results.
Lastly a gradient boosting machine is then used to make our final classification giving decent final results.
(2020)Multimodal fusion with deep neural networks for leveraging CT imaging and electronic health record.
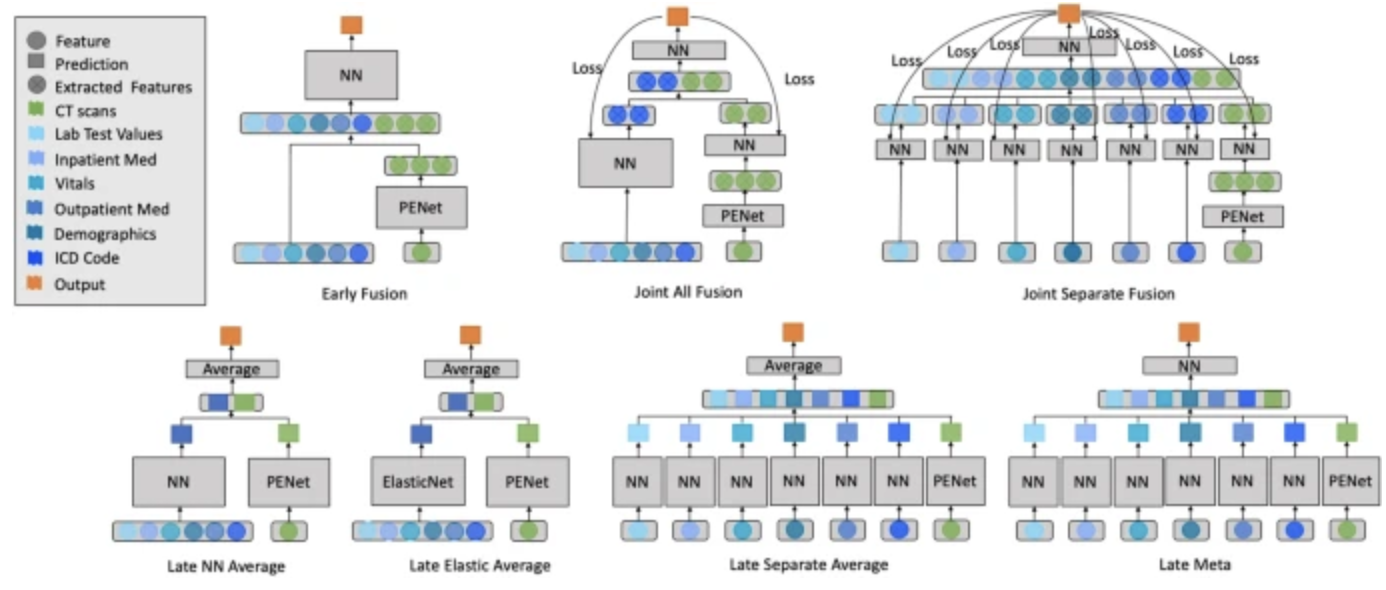
This paper is looking at how various multimodal model architectures are used to solve the diagnosis of pulmonary embolisms.
Grid search was used to find optimal hyperparameters. The late elastic average model achieved the highest AUROC of 0.947, outperforming significantly. This makes sense as a single NN can interact with each feature of the same domain and then concatenate results. It would’ve been interesting to put a NN at the end as well (that may perform better) but the results are still good!
Their late fusion model takes the averages of two separate models, meaning that even if there’s a missing modality a prediction can still be made.
(2020)[Multimodal Brain Tumor Classification Using Deep Learning and Robust Feature Selection: A Machine Learning Application for Radiologists]
This paper uses contrast stretching to stretch the minimum and maximum intensity values, as neurological imaging can often be hard to evaluate. Remember from my biomedical signal processing blog, a lot of the focus was on properly handling just this type of imaging.
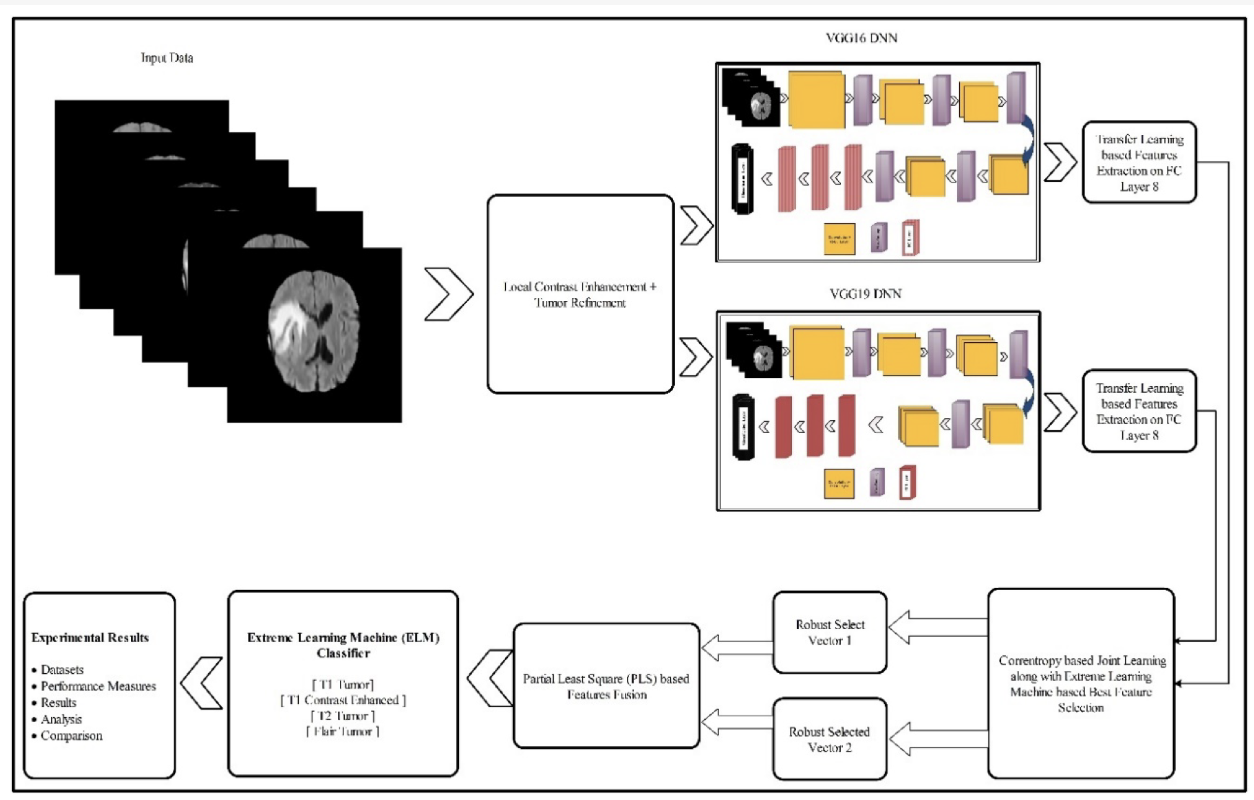
The images are fed into a pre-trained CNN VGG16 and VGG19 for feature extraction. The embedded vectors are then fed into an extreme learning machine (ELM). ELMs are structurally the same as neural networks but they do all of their optimization immediately and not iteratively using backpropagation. These are used for smaller datasets to get quick decent results that don’t have the same level of generalization as a neural network.
The features of the ELM are fed into a Partial Least Squares Regression classifier (PLS). To understand what this is we have to go from reviewing PCA, to PCR to then PLS.
PCA (principal component analysis) finds the center of the data and then sets the center to the axis of the graph. It finds a line that fits the data the best. Keep fitting free perpendicular lines and those lines make up the new plot. Rotate points according to those lines. A scree plot then finds the variation in the data according to each new partial component (PC) line. Then if we have 4 PCs and only 2 on the scree plot demonstrate importance, our 4d plot is plotted onto the 2d plot.
PCR (principal component regression) does PCA first to reduce dimensionality. We can then fit a linear regression line to this data. A normal problem in regression is that multiple variables may have similar regression lines and predictions that result in a good prediction, but only one variable is actually the cause; multicollinearity. The predictors are made harder to analyze however as they’re linear combinations of other predictors so explainability is hurt.
PLS (partial least square regression) is the same as PCR but it takes into consideration the output variable as well.
Lastly a final ELM is used to determine the results for brain tumors.
(2020)Fusion of medical imaging and EHRs using DL: a systematic review and implementation guidelines
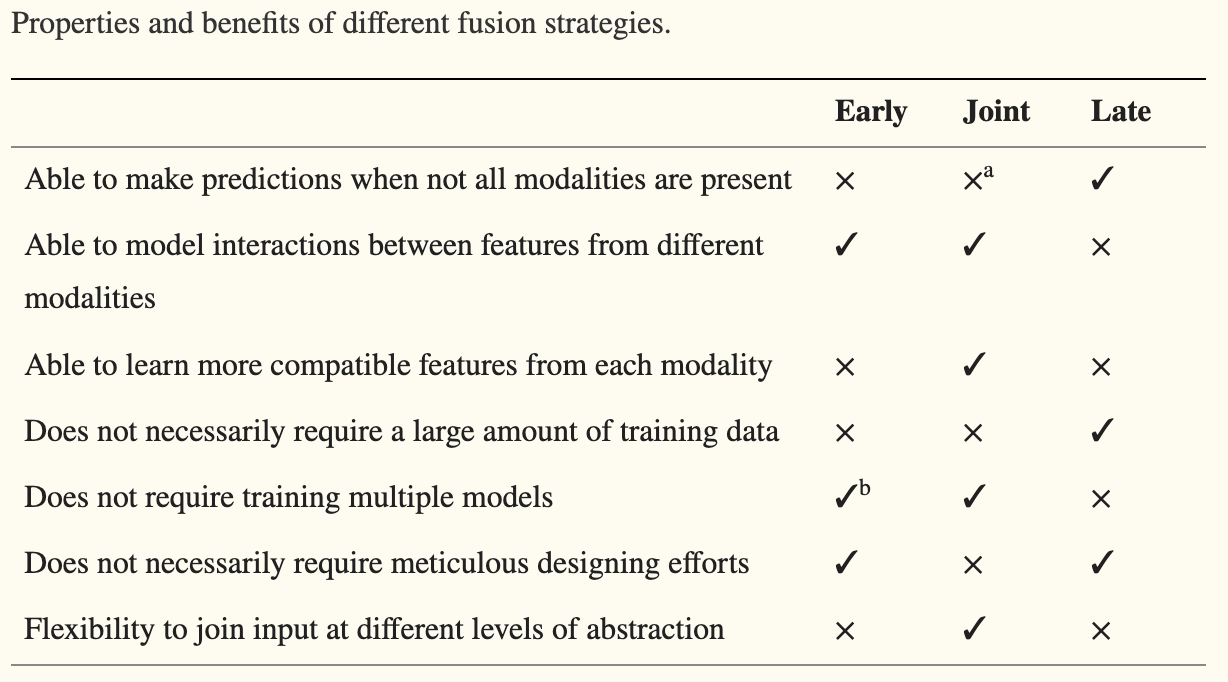
This was a great summary of 17 papers on multimodal papers with a focus on a combination of imaging and EHR data. Early fusion concatenating inputs like PET and MRI images occurred frequently, with the authors arguing for input modalities that are similar enough to be concatenated.
Join fusion already uses NNs on separate modalities and concatenates the embeddings for another layer of NNs to continue analyzing the data. Join fusion doesn’t work well with few samples relative to the number of features as there’s just too much to learn.
Late fusion uses different aggregation strategies where an ensemble method of voting is used. When modalities are independent of each other entirely, late fusion is preferred. This form of data is also good for instances of missing data, as late fusion modalities can have features excluded without a drastic dip in performance.
Fusion highly correlated features in earlier layers and less correlated features in deeper layers improves model performance. Other pros and cons are listed below.
(2021)A comprehensive survey on multimodal medical signals fusion for smart healthcare systems
With more and more electronic devices in circulation, the phrase “internet of things” (IoT) is used to describe the wide array of information that is made available to people. This can be greatly utilized for multimodal healthcare servicing.
I mostly used this paper to find other interesting papers I may have missed as everything else mentioned here was talked about before.
(2021)Mufasa: Multimodal fusion architecture search for electronic health records
Now THIS was a truly great paper. Neural Architecture Search (NAS) is the automatic design of neural networks. Everything from handling modalities to proper fusion of data, everything. NAS has normally worked for unimodal data, while Mufasa (MUltimodal Fusion Architecture SeArch) looks to remedy this. And yes, this naming convention is terrible haha.
Developing the best NAS starts by creating a known strong architecture by which we can slowly tinker with and adapt. We assign fitness scores to models, train them, and alter ones that do better and eliminate those that don’t in a “tournament selection” otherwise known as the “genetic algorithm”. This is the normal NAS strategy, except now we implement another mutation feature of concatenating different modalities at all possible states.
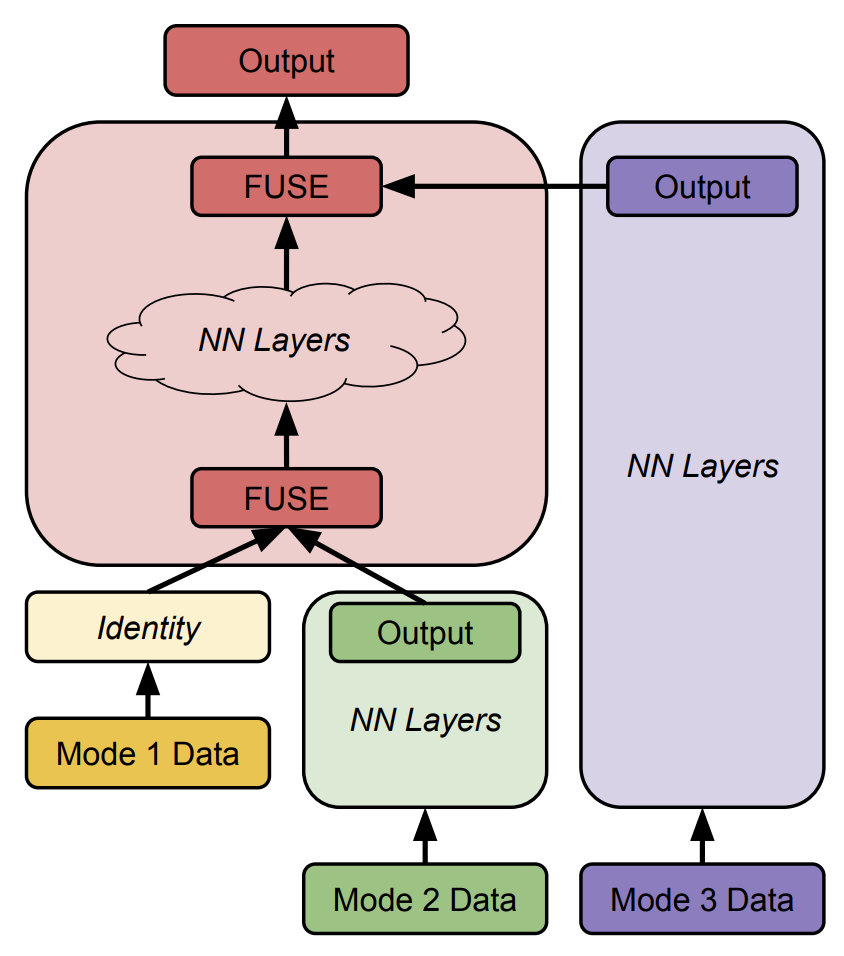
This is an example image in how modalities are fused at different levels, initially at random, but over time converge to a locally best result.
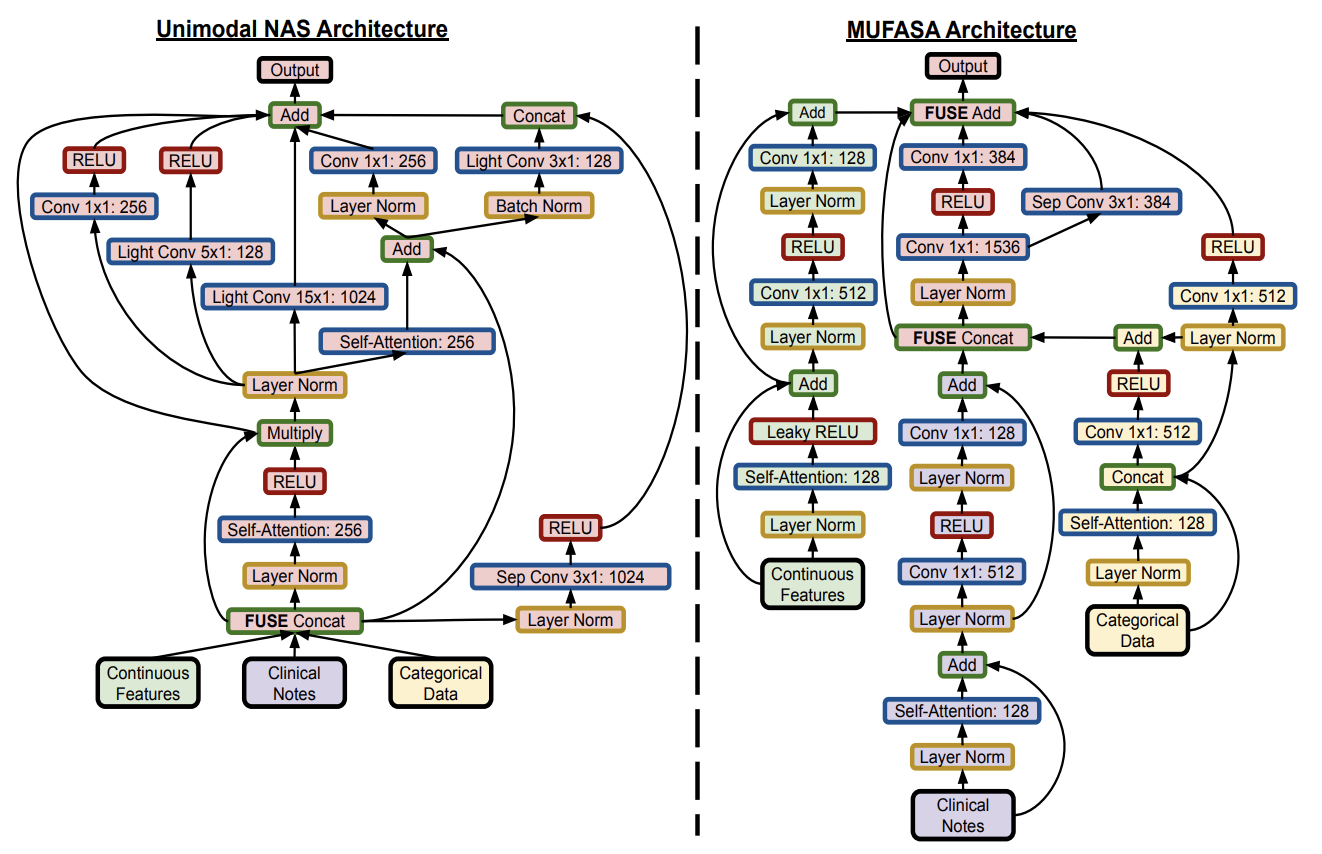
These fusions do get a bit crazy and risk overfitting and it isn’t something that’s addressed by the authors. Nonetheless this technique is incredibly valuable. It’s worth noting that previous papers like the prostate cancer and GCC couldn’t be developed by this algorithm, as specialized models will outperform any basic Mufasa NAS, however the addition of this NAS to the existing models is something that could benefit more papers down the road to edge out better results. This was a truly wonderful paper to read that really pushed the boundaries on what’s possible!
(2022)A stack based multimodal ML model for breast cancer diagnosis
This is another standard application of existing techniques to a new dataset.
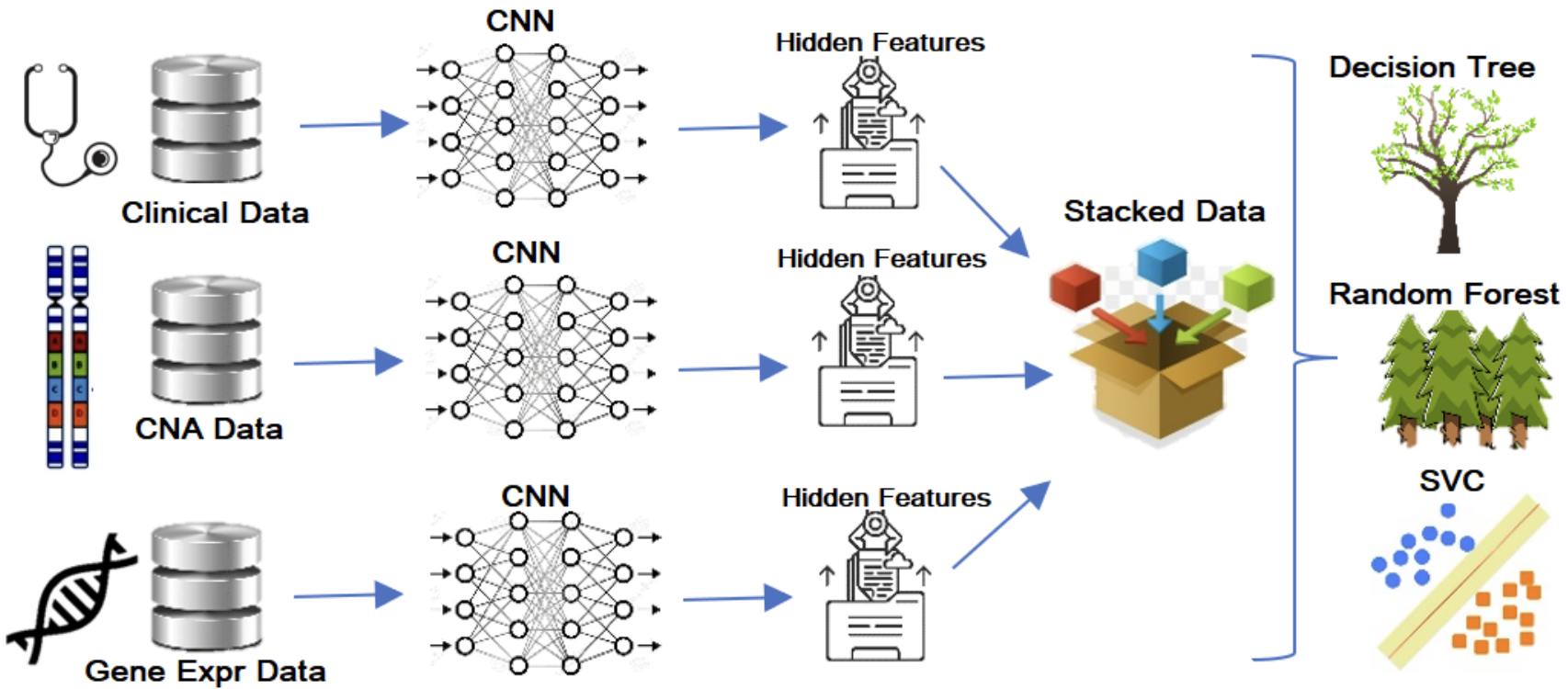
We’re taking clinical, CNA, and gene expression data, passing them all through varying CNNs and concatenating their results. The embeddings are then passed to the random forest as it was the best classifier among the three tested classifiers. With limited data they opted for low-data classifiers as shown above as well as ten-fold cross-validation.
Maximum Relevance - Minimum Redundancy (MRMR) feature selection algorithm was used to reduce the dimensionality of gene expression and CNA datasets. Every iteration the goal is to choose the most relevant property relating to the objective variable and the least redundant property that has been chosen at prior iterations.
(2022)Multimodal biomedical AI
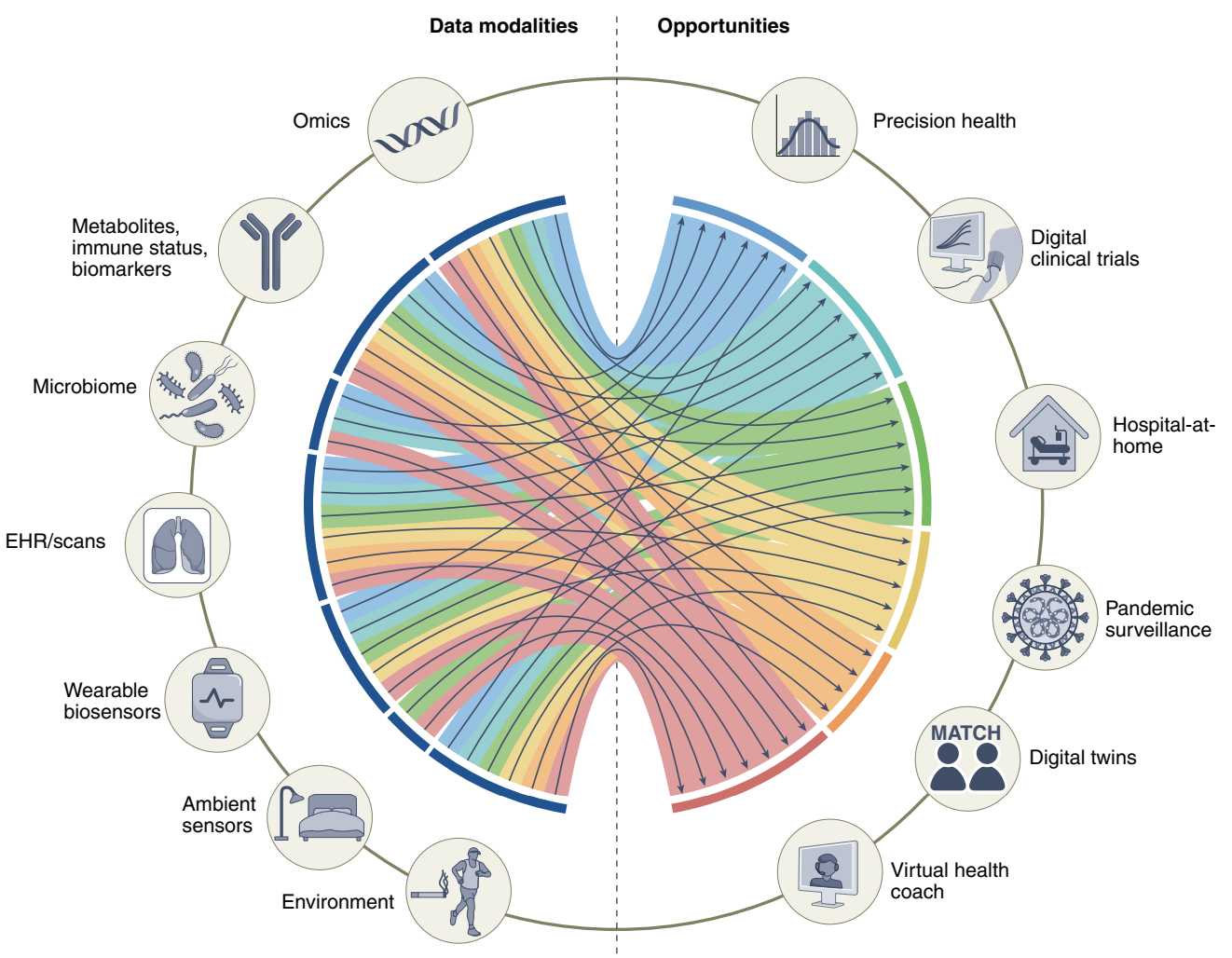
This paper starts off with a good introduction to multimodal data potentials, as well as giving a great diagram for what variables can be involved in which parts of health.
Digital twin technology is cool because the human body is complicated and many drugs (especially psychological) affect people differently. Unlearn.AI has developed and tested digital twin models that take into account various clinical data to enhance clinical trials for Alzheimer’s disease and multiple sclerosis!
Learning accurate representations for similar modalities is important, such as a picture of an apple and the word “apple” being close together. This would be referred to as Contrastive Language Image Pretraining (CLIP) originally shown by OpenAI. Contrastive Learning is a promising field that may be useful in many applications for unsupervised representation learning. This would be great for image labeling. In an embedding space images that are similar are pulled towards each other and at the same time are pushed away from differing images.
A transformer that can take in multimodal data in any form (domain-agnostic) and produce good classifications would be incredible, and Meta AI proposed a framework and a Domain-Agnostic Benchmark for Self-supervised learning (DABS) benchmark containing chest X-rays, sensor data and natural image and text data.
Alphabet’s Perceiver IO is a proposed framework for learning across modalities with the same backbone architecture. They put all information into modality-agnostic byte arrays that condense everything through an attention bottleneck to avoid large memory costs.
There was a lot more here, this was just what I was interested in and what I’ll continue to research in full if I do more multimodal data research. These technologies are super cool!
Thank you for reading! Why? Really and truly why? Unless you’re a researcher I have no clue why you put up with this, but if it was useful in any way I’m glad! I hope you have a wonderful rest of your day! :)